PEACH Lab
The purpose of this tutorial is to explain how to start working with PEACH Lab environment where users can use Jupyter notebooks to explore data and build recommendation algorithms.
Prerequisites
To be able to complete this tutorial, you will need an access to the PEACH Lab. It is required to have EBU GitLab account and permissions to use PEACH Lab. If you are new to the PEACH contact the team to get it solved for you.
Starting PEACH Lab
First you need to set up your notebook engine on PEACH platform, where you first sign in using GitLab account.
Spawner options
Before starting your PEACH Lab environment you are given a choice for some customization:

-
Cloning code repositories
PEACH Lab is integrated with EBU GitLab and it is possible to clone repositories from GitLab into your Lab environment If you want to work with your repositories in PEACH Lab - now it's a good time to select them. Before that you need to tag your repositories on GitLab with label "peach-lab" (and refresh the page to get access to the new repositories):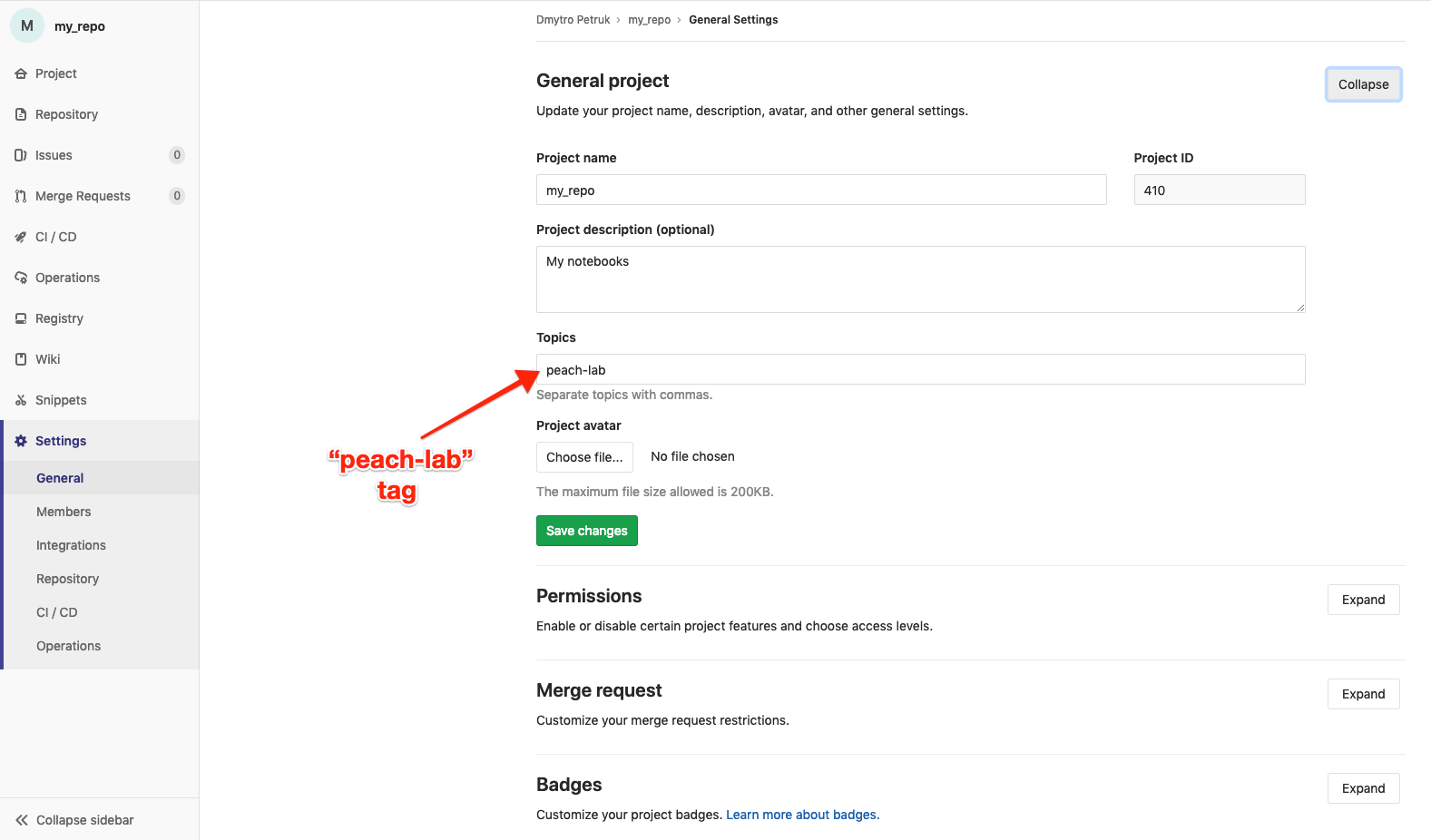
Select repositories which you would like to be cloned to your environment during the engine start. You can choose private (your personal) or public repositories (broadcaster/team level).
Preferred way to organize the code is by creating folder
notebooks/in the root folder and placing your notebooks there, creating subfolders on per project basis when such need arises -
Generation new PEACH Lab from the template
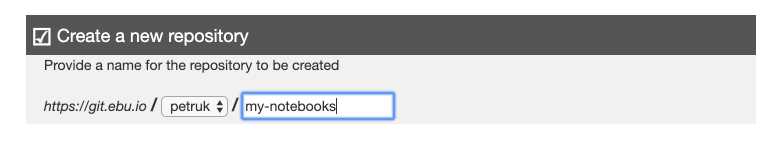
Generate new project with suggested folder structure with an already defined task and endpoint to kickstart working with the PEACH Lab platform
About the Jupyterlab
The Jupyter Notebook is an open-source web application that allows you to create and share documents that contain live code, equations, visualizations and narrative text. Uses include: data cleaning and transformation, numerical simulation, statistical modeling, data visualization, machine learning, and much more.
After spawning the environment, which may take around a minute, Jupyterlab window will have the following look:
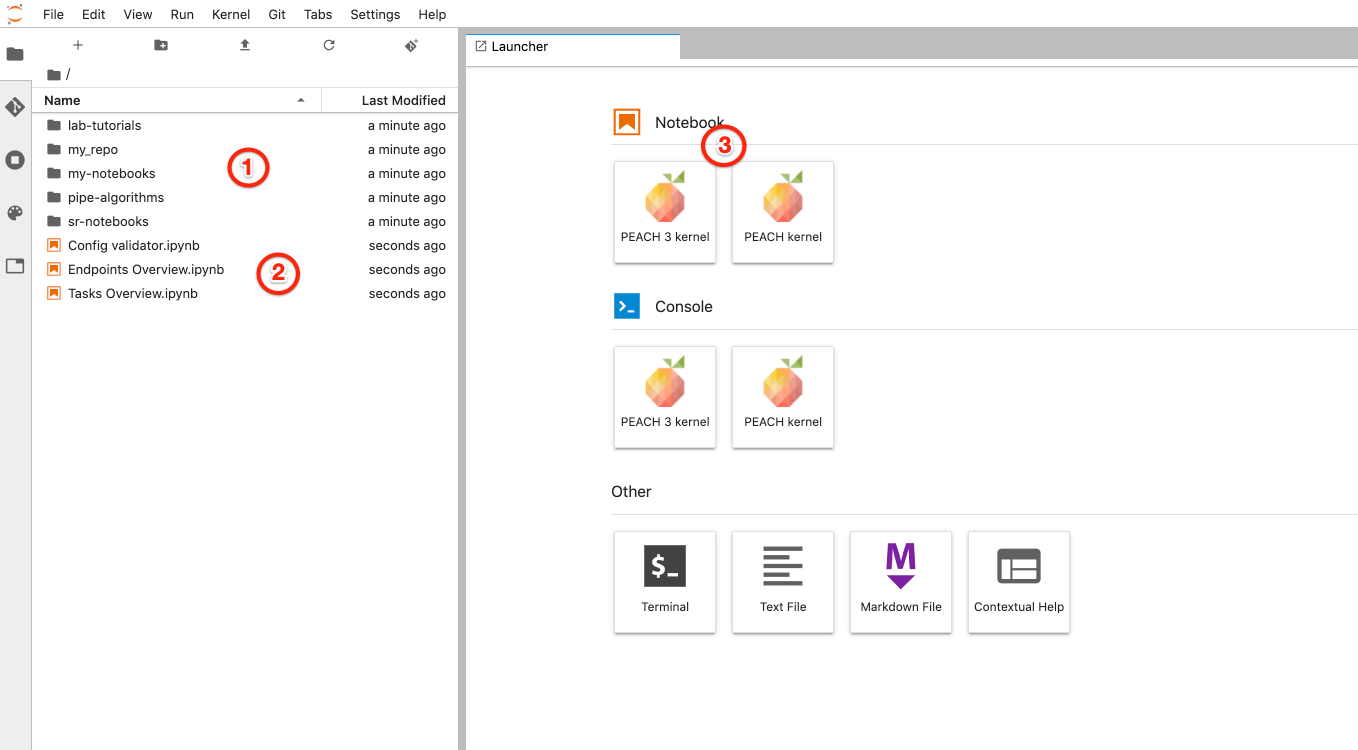
- Git repositories you have access to. Includes common libraries, repositories selected during previous step and optionally your scaffolded repository
- Overview notebooks to view status of tasks and endpoints, notebook to validate configuration files
- To start interactive Jupyter notebook session with PEACH environment, including installed dependencies with access to Redis and Spark environments.
Git integration
PEACH Lab is integrated with EBU GitLab so you can perform various operations inside git repository using UI:
- pull
- create branches
- revise changes in history
- diff notebooks
- stage changes for commit
- commit
- push
DVC integration
PEACH Lab has integration with Data Version Control system, for the purpose of sharing and versioning datasets, trained models and other large files that should not be stored in the git repository.
The idea of the DVC is to have similar interface to the Source Version Control (like git), just for large files. For that, an additional scalable storage is used (in PEACH we use AWS S3 and it's configured out-of-the-box).
Now, let's imagine we have some large file data/dataset.pkl, a fixed dataset, that we want to share with other data scientists or to train the model with. Then:
dvc add data/dataset.pkl. Create a reference to our dataset file and adds original file to .gitignoregit add data/dataset.pkl.dvc data/.gitignore. Add to git the reference to our dataset file and .gitignore (saying to ignore original file for tracking)dvc push. Push our original dataset to S3 (the process may take a while, depending on the file size)git commit -m 'Added new dataset'. Commit recent git changesgit push origin master. Push to the remote git repository
Original file now is located on S3 and if the git repository is cloned by other people/services - it will not have the dataset, only the reference to it. In order to download the dataset:
git pull origin master. This pulls reference to the datasetdvc pull. Pull the actual dataset file from S3